Protein
Huilin Shao,Jaehoon Chung, Leonora Balaj, Alain Charest, Darell D Bigner, Bob S Carter, Fred H
Hochberg, Xandra O Breakefield, Ralph Weissleder & Hakho Lee (2012) Protein typing of circulating microvesicles allows realtime monitoring of glioblastoma therapy, Nature Medicine | Technical Report, doi:10.1038/nm.2994, Published online 11 November 2012
Glioblastomas shed large quantities of small, membranebound microvesicles into the circulation. Although these hold promise as potential biomarkers of therapeutic response, their identification and quantification remain challenging. Here, we describe a highly sensitive and rapid analytical technique for profiling circulating microvesicles directly from blood samples of patients with glioblastoma. Microvesicles, introduced onto a dedicated microfluidic chip, are labeled with targetspecific magnetic nanoparticles and detected by a miniaturized nuclear magnetic resonance system. Compared with current methods, this integrated system has a much higher detection sensitivity and can differentiate glioblastoma multiforme (GBM) microvesicles from nontumor host cell–derived microvesicles. We also show that circulating GBM microvesicles can be used to analyze primary tumor mutations and as a predictive metric of treatmentinduced changes. This platform could provide both an early indicator of drug efficacy and a potential molecular stratifier for human clinical trials.
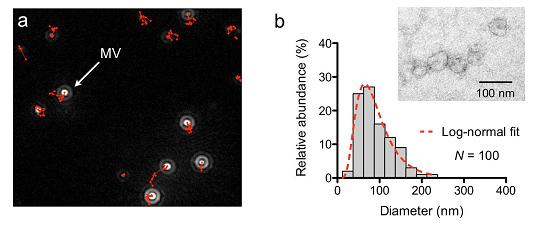
Supplementary Figure 1. Microscopy analysis of MVs. (a) MVs from GBM20/3 primary human glioblastoma cell line, previously used in other MV studies6,7, were counted by nanoparticle tracking analysis (NTA). The system monitored the trajectory of MV movement (in red) to obtain MV sizes and concentrations. NTA was initially used to estimate the MV concentration in a given sample. For consistent reading, the measurement settings were optimized and maintained for a .detection range of 10 500 nm. (b) Size distribution of MVs purified by the developed microfluidic device.
Purified GBM20/3 MVs were imaged by transmission electron microscopy (inset), and thesize histogram was obtained by measuring 100 MVs. The prepared MVs have a lognormal sizedistribution (dotted line) with a median diameter of 83 nm. Note that about 75% of the prepared MV population was less than 120 nm in diameter.